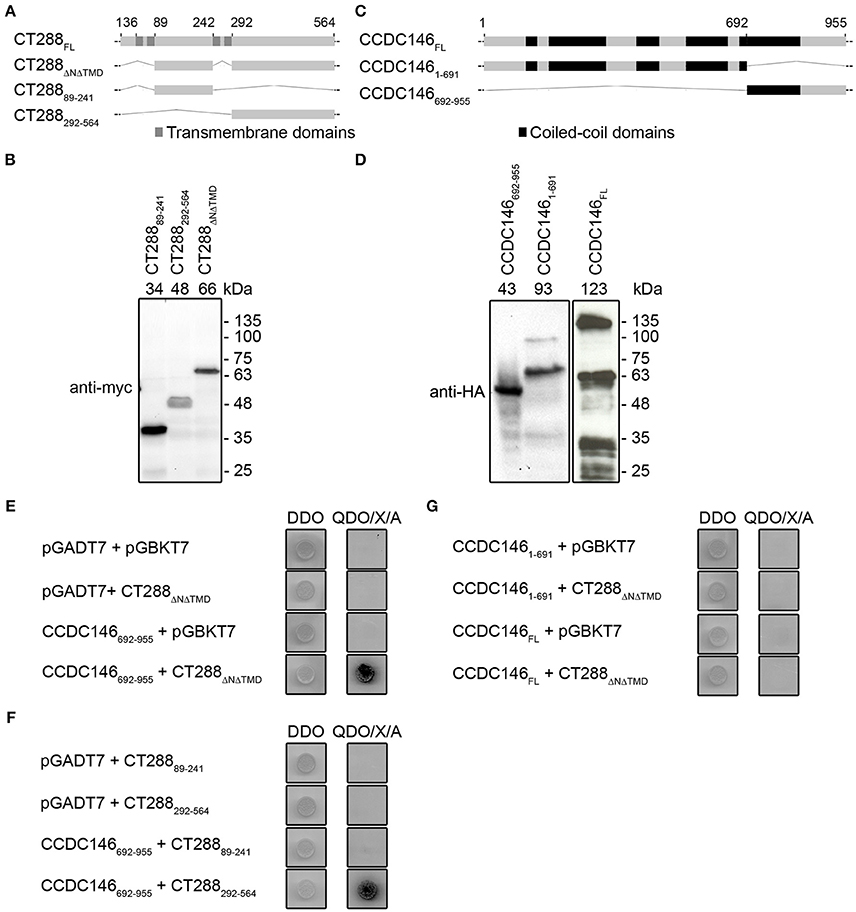
Frontiers | The Human Centrosomal Protein CCDC146 Binds Chlamydia trachomatis Inclusion Membrane Protein CT288 and Is Recruited to the Periphery of the Chlamydia-Containing Vacuole

Chlamydia trachomatis Inclusion Membrane Protein CT228 Recruits Elements of the Myosin Phosphatase Pathway to Regulate Release Mechanisms - ScienceDirect

Chloramphenicol (Cam) affects chlamydial growth and GFP expression. (a)... | Download Scientific Diagram
The Chlamydia trachomatis inclusion membrane protein CT006 associates with lipid droplets in eukaryotic cells | PLOS ONE
Quantitative Monitoring of the Chlamydia trachomatis Developmental Cycle Using GFP-Expressing Bacteria, Microscopy and Flow Cytometry | PLOS ONE
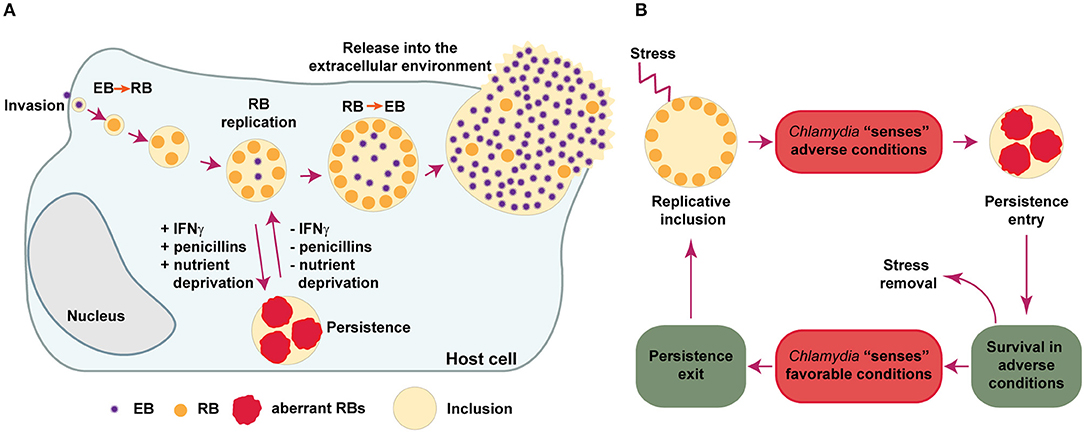
Frontiers | Chlamydia Persistence: A Survival Strategy to Evade Antimicrobial Effects in-vitro and in-vivo

Cell Type Development in Chlamydia trachomatis Follows a Program Intrinsic to the Reticulate Body | bioRxiv
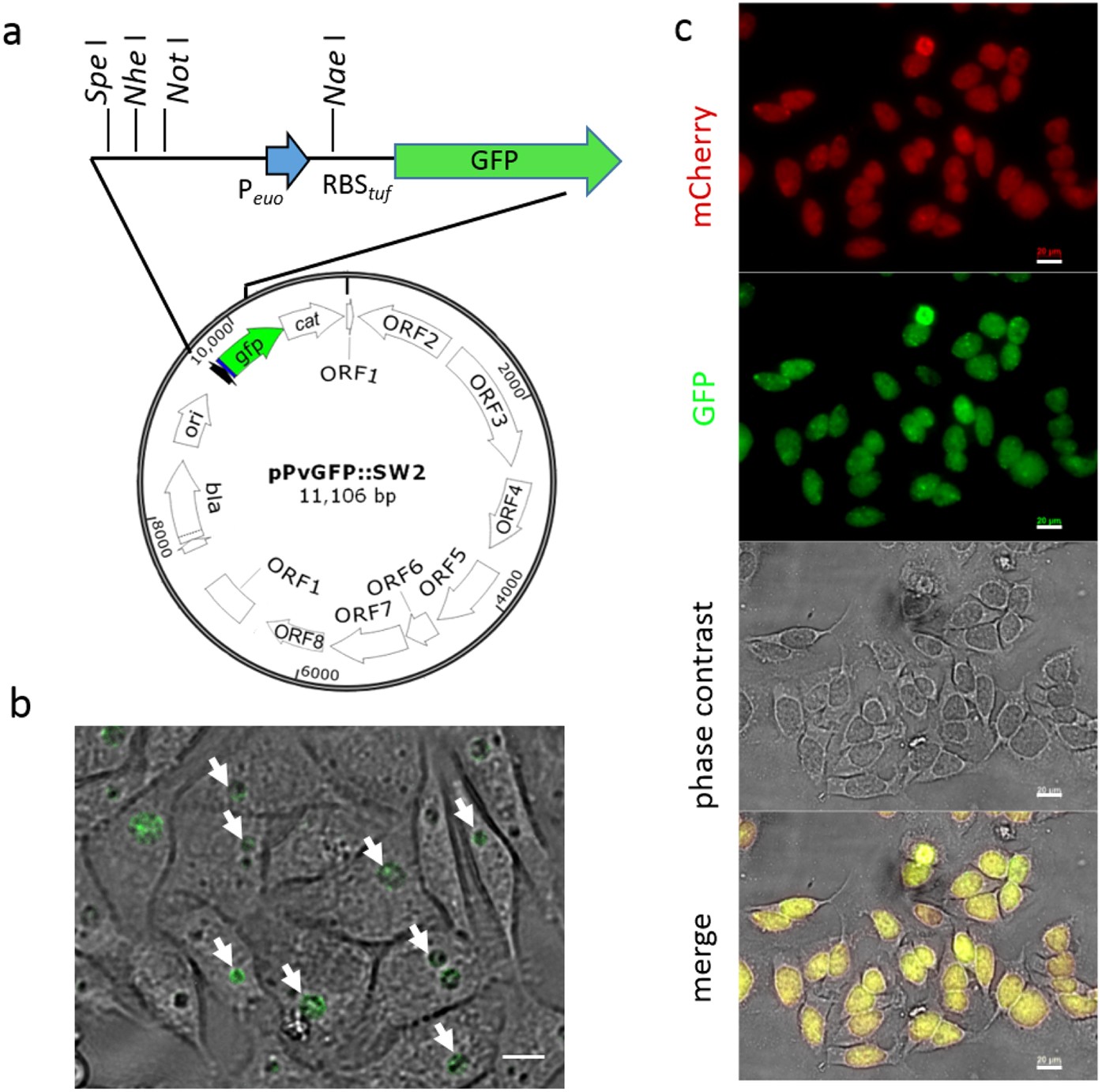
Quantifying promoter activity during the developmental cycle of Chlamydia trachomatis | Scientific Reports

Detailed visualization of the Chlamydia developmental cycle in GFP-HeLa... | Download Scientific Diagram
The Chlamydia trachomatis inclusion membrane protein CT006 associates with lipid droplets in eukaryotic cells | PLOS ONE

Chlamydia trachomatis Encodes a Dynamic, Ring-Forming Bactofilin Critical for Maintaining Cell Size and Shape | bioRxiv
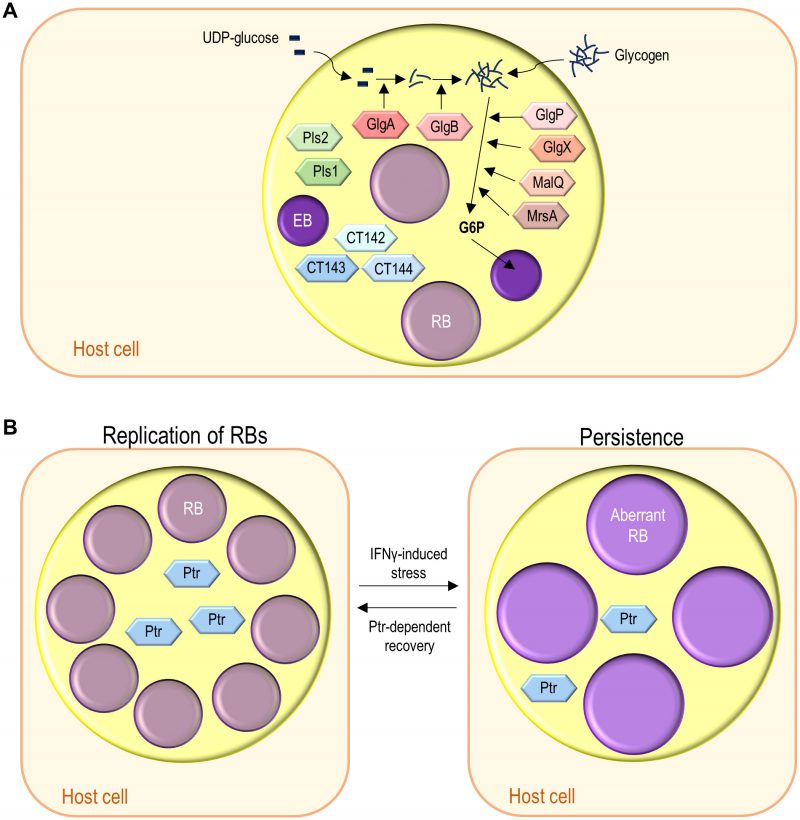
The multiple functions of the numerous Chlamydia trachomatis secreted proteins: the tip of the iceberg
Pathogenic Chlamydia Lack a Classical Sacculus but Synthesize a Narrow, Mid-cell Peptidoglycan Ring, Regulated by MreB, for Cell Division | PLOS Pathogens

The Chlamydia trachomatis Inclusion Membrane Protein CpoS Counteracts STING-Mediated Cellular Surveillance and Suicide Programs - ScienceDirect
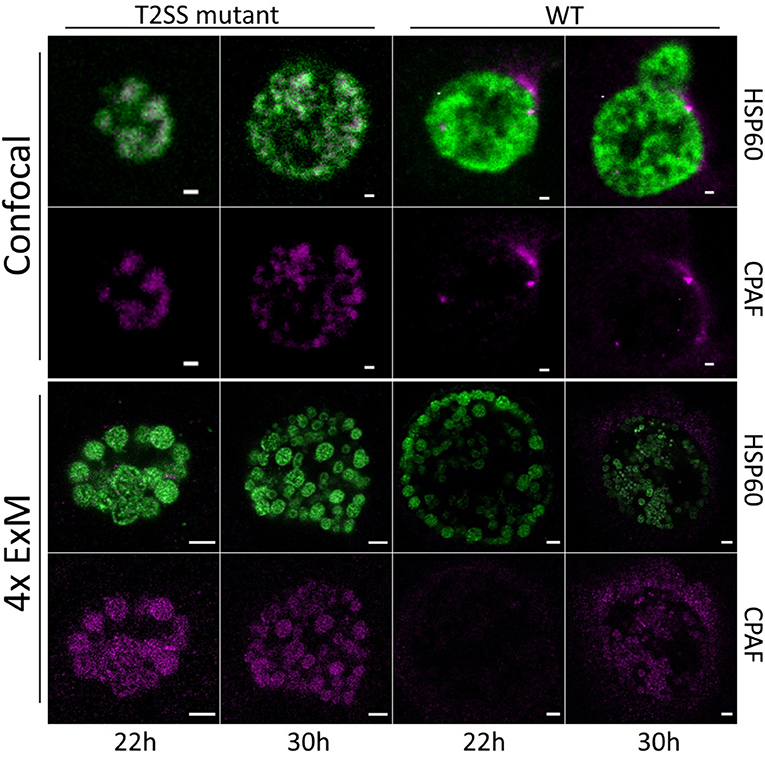
Frontiers | Detection of Chlamydia Developmental Forms and Secreted Effectors by Expansion Microscopy

The Chlamydia protein CpoS modulates the inclusion microenvironment and restricts the interferon response by acting on Rab35 | bioRxiv
The Chlamydia trachomatis inclusion membrane protein CT006 associates with lipid droplets in eukaryotic cells | PLOS ONE

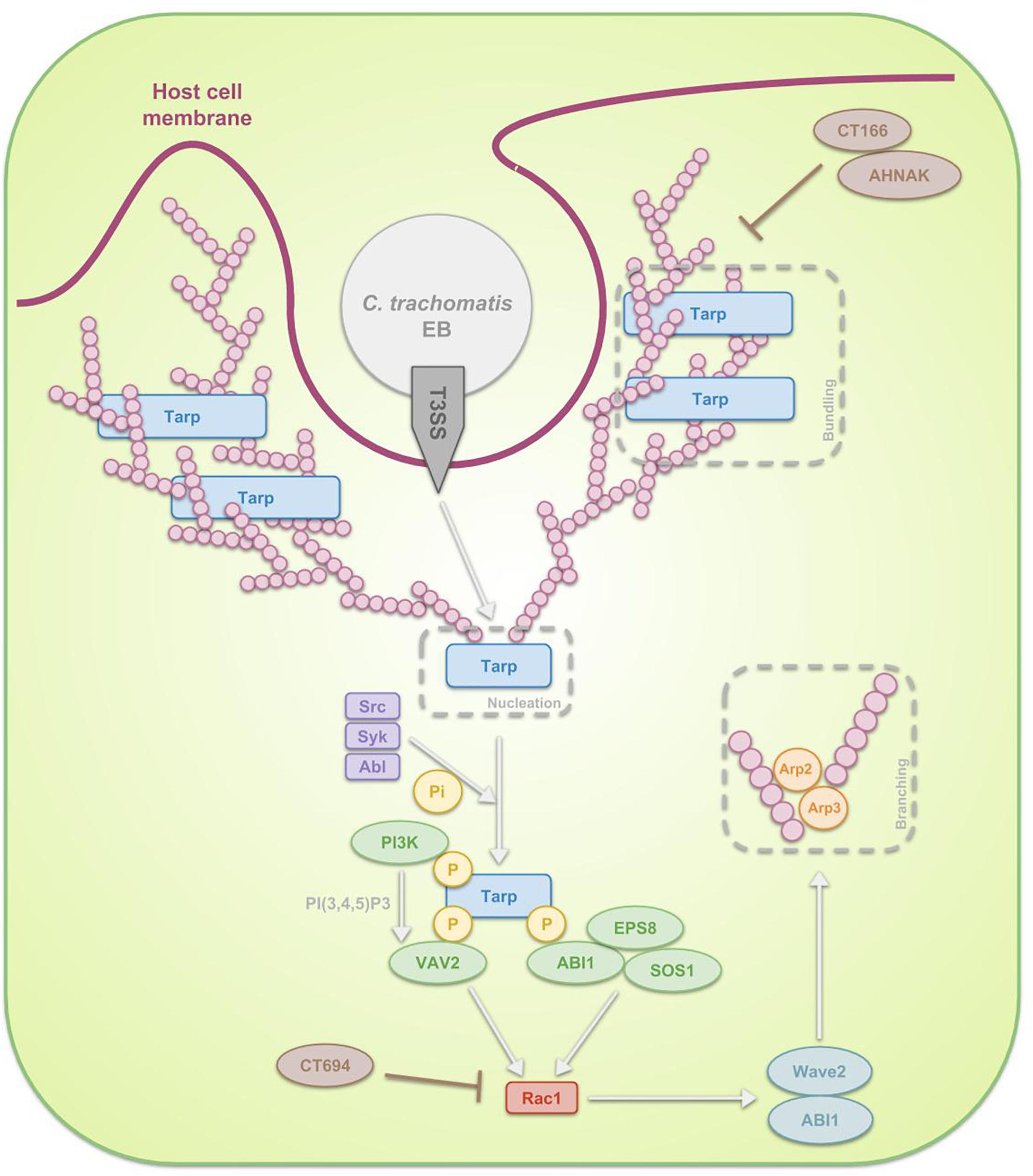
Frontiers | The Chlamydia trachomatis Early Effector Tarp Outcompetes Fascin in Forming F-Actin Bundles In Vivo
The Chlamydia trachomatis inclusion membrane protein CT006 associates with lipid droplets in eukaryotic cells | PLOS ONE
Chlamydia trachomatis and Chlamydia muridarum spectinomycin resistant vectors and a transcriptional fluorescent reporter to monitor conversion from replicative to infectious bacteria | PLOS ONE
Proximity-dependent proteomics of the Chlamydia trachomatis inclusion membrane reveals functional interactions with endoplasmic reticulum exit sites | PLOS Pathogens